Topology of viral evolution
Joseph M Chan, Gunnar Carlsson, Raul Rabadan. Topology of viral evolution. PNAS, 2013: 110(46), 18566-18571. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3831954/
Significance
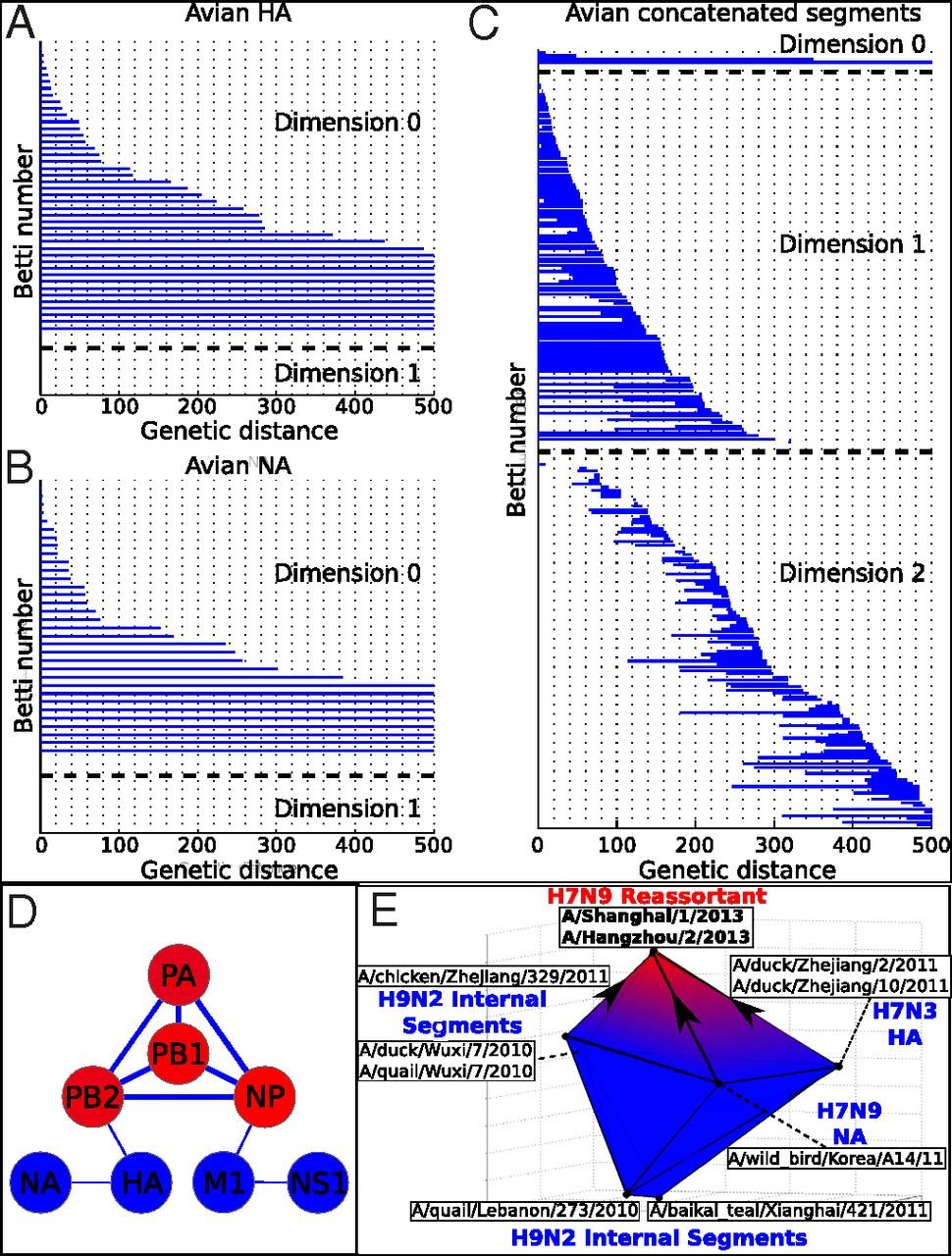
Evolution is mediated not only by random mutations over a number of generations (vertical evolution), but also through the mixture of genomic material between individuals of different lineages (horizontal evolution). The standard evolutionary representation, the phylogenetic tree, faithfully represents the former but not the latter scenario. Although many elaborations have been developed to address this issue, there is still no agreed-upon method of incorporating both vertical and horizontal evolution. Here, we present an alternative strategy based on algebraic topology to study evolution. This method extends beyond the limits of a tree to capture directly even complex horizontal exchanges between multiple parental strains, as well as uncover broader reticulate patterns, including the segregation of segments during reassortment.
Abstract
The tree structure is currently the accepted paradigm to represent evolutionary relationships between organisms, species or other taxa. However, horizontal, or reticulate, genomic exchanges are pervasive in nature and confound characterization of phylogenetic trees. Drawing from algebraic topology, we present a unique evolutionary framework that comprehensively captures both clonal and reticulate evolution. We show that whereas clonal evolution can be summarized as a tree, reticulate evolution exhibits nontrivial topology of dimension greater than zero. Our method effectively characterizes clonal evolution, reassortment, and recombination in RNA viruses. Beyond detecting reticulate evolution, we succinctly recapitulate the history of complex genetic exchanges involving more than two parental strains, such as the triple reassortment of H7N9 avian influenza and the formation of circulating HIV-1 recombinants. In addition, we identify recurrent, large-scale patterns of reticulate evolution, including frequent PB2-PB1-PA-NP cosegregation during avian influenza reassortment. Finally, we bound the rate of reticulate events (i.e., 20 reassortments per year in avian influenza). Our method provides an evolutionary perspective that not only captures reticulate events precluding phylogeny, but also indicates the evolutionary scales where phylogenetic inference could be accurate.